Image-Based Modeling and Analysis of Cardiac Microbundles
Outcome/Accomplishment
Researchers at the U.S. National Science Foundation (NSF)-funded Cellular Metamaterials (NSF CELL-MET) Engineering Research Center (ERC) based at Boston University (BU) and the University of Michigan, Ann Arbor (UM-AA) introduced a computational modeling and analysis pipeline for cardiac microbundles and their contractions. Additionally, the researchers developed and published MicrobundleCompute software, which offers tools for automatic tissue segmentation, tracking, and analysis of videos of beating cardiac microbundles.
Impact/Benefits
A core challenge for advances such as disease modeling, drug discovery, and regenerative tissue engineering is the need to attain mature human-induced pluripotent stem-cell derived cardiomyocyte (hiPSC-CM) tissues. However, limited options have been available for producing reliable and reproducible functional metrics that properly serve cross-system needs. At the same time, there is opportunity to better leverage information available in time-lapse images of cardiac microbundle contractions. The computational framework established by NSF CELL-MET enables automatic quantification of morphology-based mechanical metrics from videos of cardiac microbundles. The modeling available via MicroBundleCompute captures the matrix and myofibrillar structure as well as functional parameters estimated based on tissue-specific data.
Explanation/Background
The computational framework offers tools for automatic tissue segmentation, tracking, and analysis of brightfield and phase contrast videos of beating cardiac microbundles. The NSF CELL-MET team describes the tools as “straightforward to implement, require[ing] little to no parameter tuning, and [capable of running] quickly on a personal computer.” Two papers were published as a result of this work. The computational tool and dataset are available in an open source software format with the aims of broader dissemination and community accessibility.
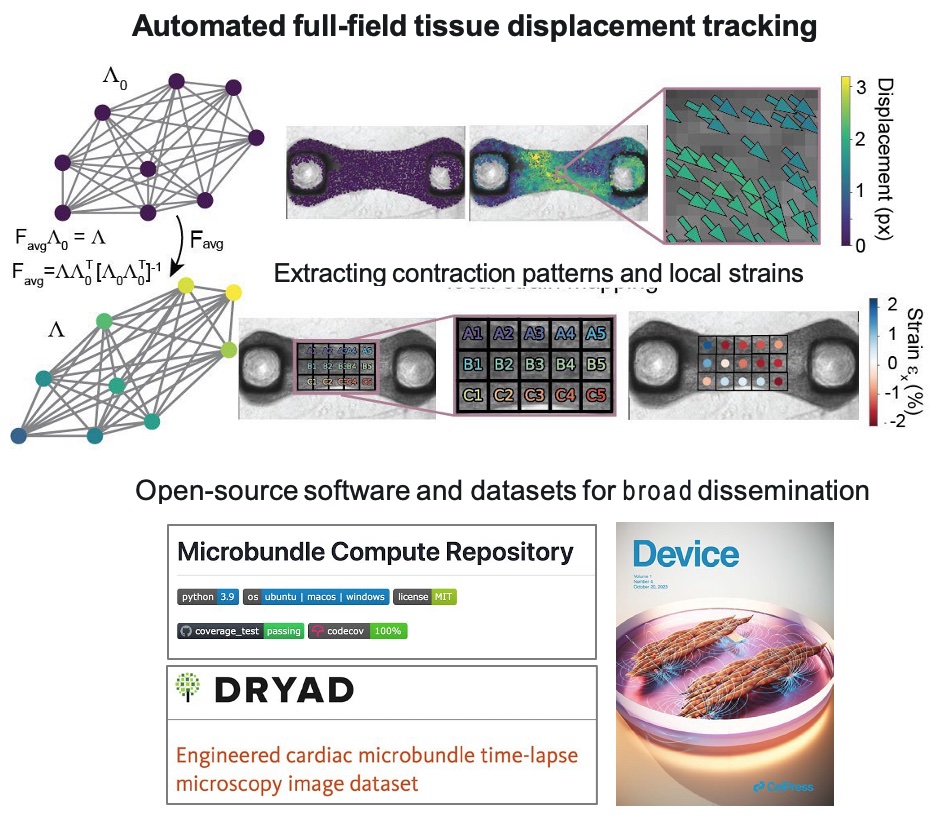
The research team led by Hiba Kobeissi at BU developed the computational framework, completed extensive validation studies, and then demonstrated use cases for functional metrics. The team automated full-field tissue displacement tracking and then showed extracting contraction patterns and local strains. A data-driven computational model for cardiac microbundles validated tissue specific models by image tracking and simulation, exploring both fiber structure and myofibril structure.
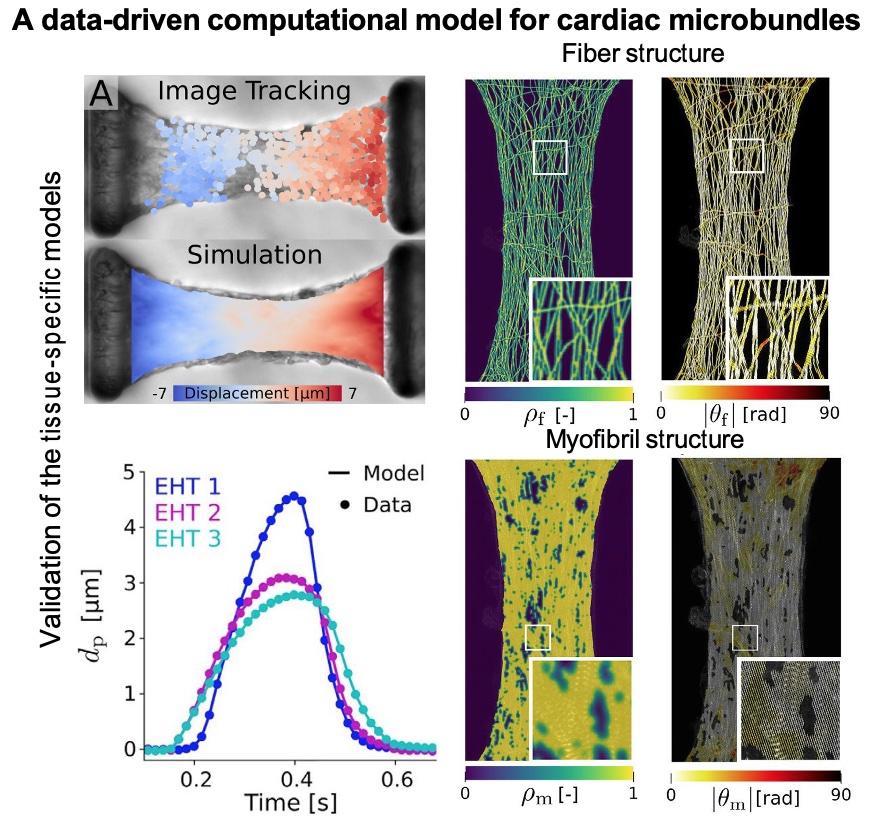
Javiera Jilberto and others at the UM-AA leveraged the new modeling pipeline to generate simulated, image-based engineered heart tissues (EHTs), capturing ECM and myofibrillar structure as well as functional parameters estimated directly from experimental data. The resulting images make it possible to estimate both EHT function as well as CM active stresses. Different mechanical environments were examined using the modeling pipeline at different CM active stress levels. The results showed that myofibril formation impacted differences in measured experimental forces; active stress appeared variably muted across conditions. The model makes it possible to experimentally understand the contributions of myofibrils as well as the extracellular matrix to tissue force output.
Location
Boston, MassachusettsStart Year
Biotechnology and Healthcare
Biotechnology and Healthcare
Lead Institution
Core Partners
Fact Sheet
Outcome/Accomplishment
Researchers at the U.S. National Science Foundation (NSF)-funded Cellular Metamaterials (NSF CELL-MET) Engineering Research Center (ERC) based at Boston University (BU) and the University of Michigan, Ann Arbor (UM-AA) introduced a computational modeling and analysis pipeline for cardiac microbundles and their contractions. Additionally, the researchers developed and published MicrobundleCompute software, which offers tools for automatic tissue segmentation, tracking, and analysis of videos of beating cardiac microbundles.
Location
Boston, MassachusettsStart Year
Biotechnology and Healthcare
Biotechnology and Healthcare
Lead Institution
Core Partners
Fact Sheet
Impact/benefits
A core challenge for advances such as disease modeling, drug discovery, and regenerative tissue engineering is the need to attain mature human-induced pluripotent stem-cell derived cardiomyocyte (hiPSC-CM) tissues. However, limited options have been available for producing reliable and reproducible functional metrics that properly serve cross-system needs. At the same time, there is opportunity to better leverage information available in time-lapse images of cardiac microbundle contractions. The computational framework established by NSF CELL-MET enables automatic quantification of morphology-based mechanical metrics from videos of cardiac microbundles. The modeling available via MicroBundleCompute captures the matrix and myofibrillar structure as well as functional parameters estimated based on tissue-specific data.
Explanation/Background
The computational framework offers tools for automatic tissue segmentation, tracking, and analysis of brightfield and phase contrast videos of beating cardiac microbundles. The NSF CELL-MET team describes the tools as “straightforward to implement, require[ing] little to no parameter tuning, and [capable of running] quickly on a personal computer.” Two papers were published as a result of this work. The computational tool and dataset are available in an open source software format with the aims of broader dissemination and community accessibility.
The research team led by Hiba Kobeissi at BU developed the computational framework, completed extensive validation studies, and then demonstrated use cases for functional metrics. The team automated full-field tissue displacement tracking and then showed extracting contraction patterns and local strains. A data-driven computational model for cardiac microbundles validated tissue specific models by image tracking and simulation, exploring both fiber structure and myofibril structure.
Javiera Jilberto and others at the UM-AA leveraged the new modeling pipeline to generate simulated, image-based engineered heart tissues (EHTs), capturing ECM and myofibrillar structure as well as functional parameters estimated directly from experimental data. The resulting images make it possible to estimate both EHT function as well as CM active stresses. Different mechanical environments were examined using the modeling pipeline at different CM active stress levels. The results showed that myofibril formation impacted differences in measured experimental forces; active stress appeared variably muted across conditions. The model makes it possible to experimentally understand the contributions of myofibrils as well as the extracellular matrix to tissue force output.